Description
Transfection reagent features:
- One single reagent is able to transfect large plasmid, mRNA, siRNA, and/or other type of nucleic acids, which is best for co-transfection of different type and/or size of nucleic acids.
- Exceptional transfection efficiency in the broadest range of cell types including cell lines of stem cell origin, suspension cells, and primary cells with less than half the amount required by competitors® products.
- Extremely gentle to cells: The chemical features of our proprietary formula and the least amount needed for transfection ensure its lowest toxicity.
- Economical: High efficiency means less amount of nucleic acid & reagent is needed. 1.5 ml of Avalanche®-Omni Reagent is sufficient for more than 3,500 transfections in 24-well plates.
- Superior performance for co-transfection of siRNA and plasmid DNA
- Proven efficacy in the presence of serum: eliminates the need to change media following transfection
- Animal free
- Reliable performance for high-throughput applications
- The best choice for establishing stable cell lines
- Simplest protocol: simply mix with nucleic acid and add to cell culture.
Data
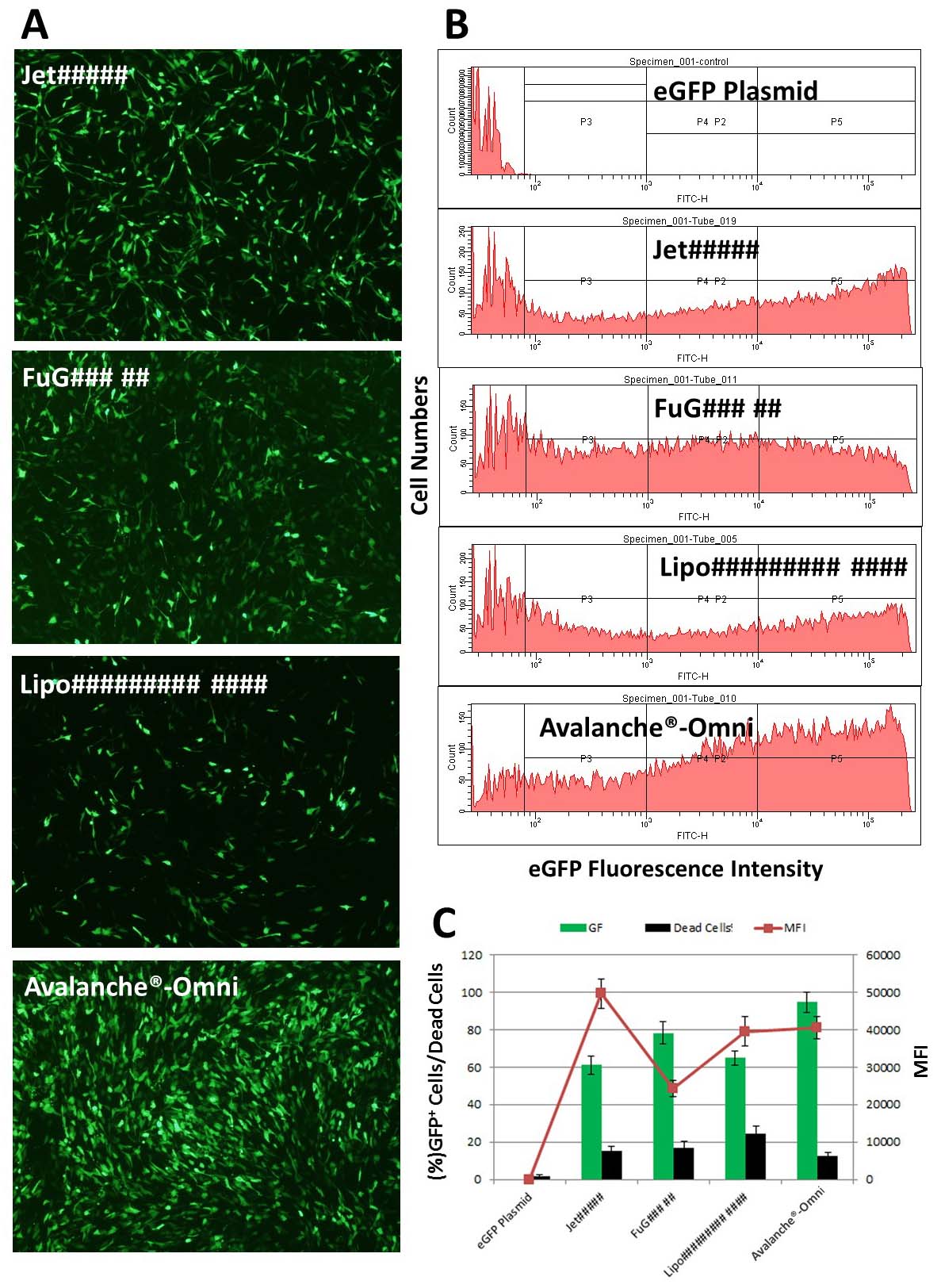 Figure 1. Avalanche®-Omni Transfection Reagent revealed highest efficiency and optimal balance of potency & low cytotoxicity. NIH 3T3 cells were transfected with enhanced green fluorescent protein (eGFP) expressing vector by using our Avalanche®-Omni Transfection Reagent and several most popular commercial transfection reagents. The transfection result was evaluated by FACS using three criteria: the percent of GFP positive cells (transfection efficiency), the percent of dead cells (toxicity, 7AAD positive cells), and mean fluorescence intensity (MFI, expression level of GFP in cells).(A) Pictures of GFP expression in cells transfected with Avalanche®-Omni and other commercial transfection reagents. (B) FACS data showed the diffeences on transfection efficiency between Avalanche®-Omni and other commercial transfection reagents. (C) Quantitative data of (B) FACS analysis: Avalanche®-Omni showed the highest transfection efficiency and the optimal balance of potent & low cytotoxicity. GFP expression level in cells are high as well in Avalanche®-Omni Transfection Reagent group.
Figure 1. Avalanche®-Omni Transfection Reagent revealed highest efficiency and optimal balance of potency & low cytotoxicity. NIH 3T3 cells were transfected with enhanced green fluorescent protein (eGFP) expressing vector by using our Avalanche®-Omni Transfection Reagent and several most popular commercial transfection reagents. The transfection result was evaluated by FACS using three criteria: the percent of GFP positive cells (transfection efficiency), the percent of dead cells (toxicity, 7AAD positive cells), and mean fluorescence intensity (MFI, expression level of GFP in cells).(A) Pictures of GFP expression in cells transfected with Avalanche®-Omni and other commercial transfection reagents. (B) FACS data showed the diffeences on transfection efficiency between Avalanche®-Omni and other commercial transfection reagents. (C) Quantitative data of (B) FACS analysis: Avalanche®-Omni showed the highest transfection efficiency and the optimal balance of potent & low cytotoxicity. GFP expression level in cells are high as well in Avalanche®-Omni Transfection Reagent group.
Avalanche®-Omni Transfection Reagent is able to achieve more than 90% knockdown of endogenous gene expression in a variety of cell lines and primary cells.
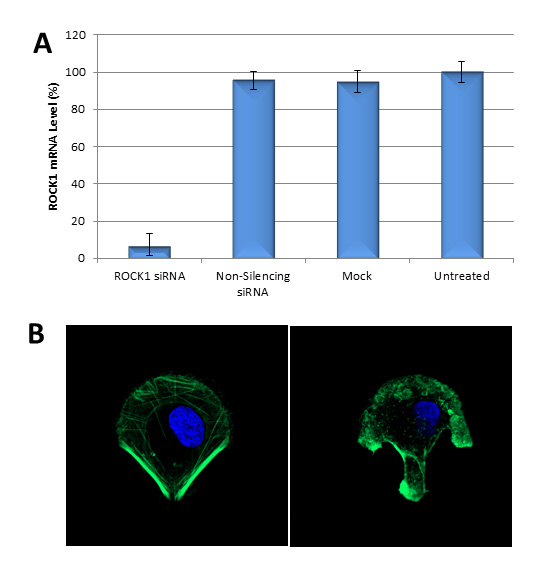 Figure 2. Avalanche®-Omni Transfection Reagent performed more than 90% knockdown of endogenous gene expression. (A). ROCK1 mRNA levels were quantified using qPCR in Hela cells transfected with Target-specific siRNA duplexes (10nM) for human ROCK1 gene or non-silencing siRNA by using Avalanche®-Omni Transfection Reagent. Data were normalized against the 18S rRNA signal. Control Samples were either mock-transected or untreated. Values are normalized to untreated sample. Data are means®SD (n=3). (B) Actin stress fibers were stained with FITC-labelled phalloidin on the cells in (A) cultured on fibronectin-coated micropattern. Confocal Microscope revealed that the one transfected with ROCK1 siRNA (right) showed disrupted stress fiber pattern as compared to the one transfected with non-silencing siRNA (left).
Figure 2. Avalanche®-Omni Transfection Reagent performed more than 90% knockdown of endogenous gene expression. (A). ROCK1 mRNA levels were quantified using qPCR in Hela cells transfected with Target-specific siRNA duplexes (10nM) for human ROCK1 gene or non-silencing siRNA by using Avalanche®-Omni Transfection Reagent. Data were normalized against the 18S rRNA signal. Control Samples were either mock-transected or untreated. Values are normalized to untreated sample. Data are means®SD (n=3). (B) Actin stress fibers were stained with FITC-labelled phalloidin on the cells in (A) cultured on fibronectin-coated micropattern. Confocal Microscope revealed that the one transfected with ROCK1 siRNA (right) showed disrupted stress fiber pattern as compared to the one transfected with non-silencing siRNA (left).
Additional Information
| Weight | 0.5 lbs |
|---|---|
| Product Sizes | 0.75 ml, 1.5 ml, 5 x 1.5 ml |
| Subcategories | Broad Spectrum |
Documents
Protocols
MSDS
Citations or Feedback
- Chaffey, J. R. (2020). Depletion of the phosphatase inhibitor, PPP1R1A, may contribute to beta-cell loss in type 1 diabetes (Doctoral dissertation, University of Exeter, United Kingdom). ProQuest Dissertations & Theses. https://www.proquest.com/dissertations-theses/30397876


