Description
Features:
- One single reagent is able to transfect large plasmid, mRNA, siRNA, and/or other type of nucleic acids, which is best for co-transfection of different type and/or size of nucleic acids.
- Broad Spectrum DNA/siRNA delivery – one transfection reagent and protocol for a variety of cells.
- Transfection efficiency is close to that of Avalanche®-Omni Transfection Reagent (EZT-OMNI-1), but with much lower prices. Its price is only 1/3 that of L2K, and 1/3.4 that of Lipofectamine® 3000 (L3K).
- Highest value:® It is able to do 3000 x 24-well transfections/1.5 ml as compared to 750-1000 x 24-well transfections/1.5 ml for L2K
- Very low Cellular Toxicity because of its bio-degradability after endocytosis. Maintain cell density, reduce experimental biases.
- Same simple protocol as that of EZT-OMNI-1: Does not require removal of serum or culture medium and does not require washing or changing of® medium after introducing the reagent/DNA complex.
- High levels of recombinant protein production
- Ideal for high-throughput work
- A must-have transfection reagent for everyday use on commonly used cells
Data
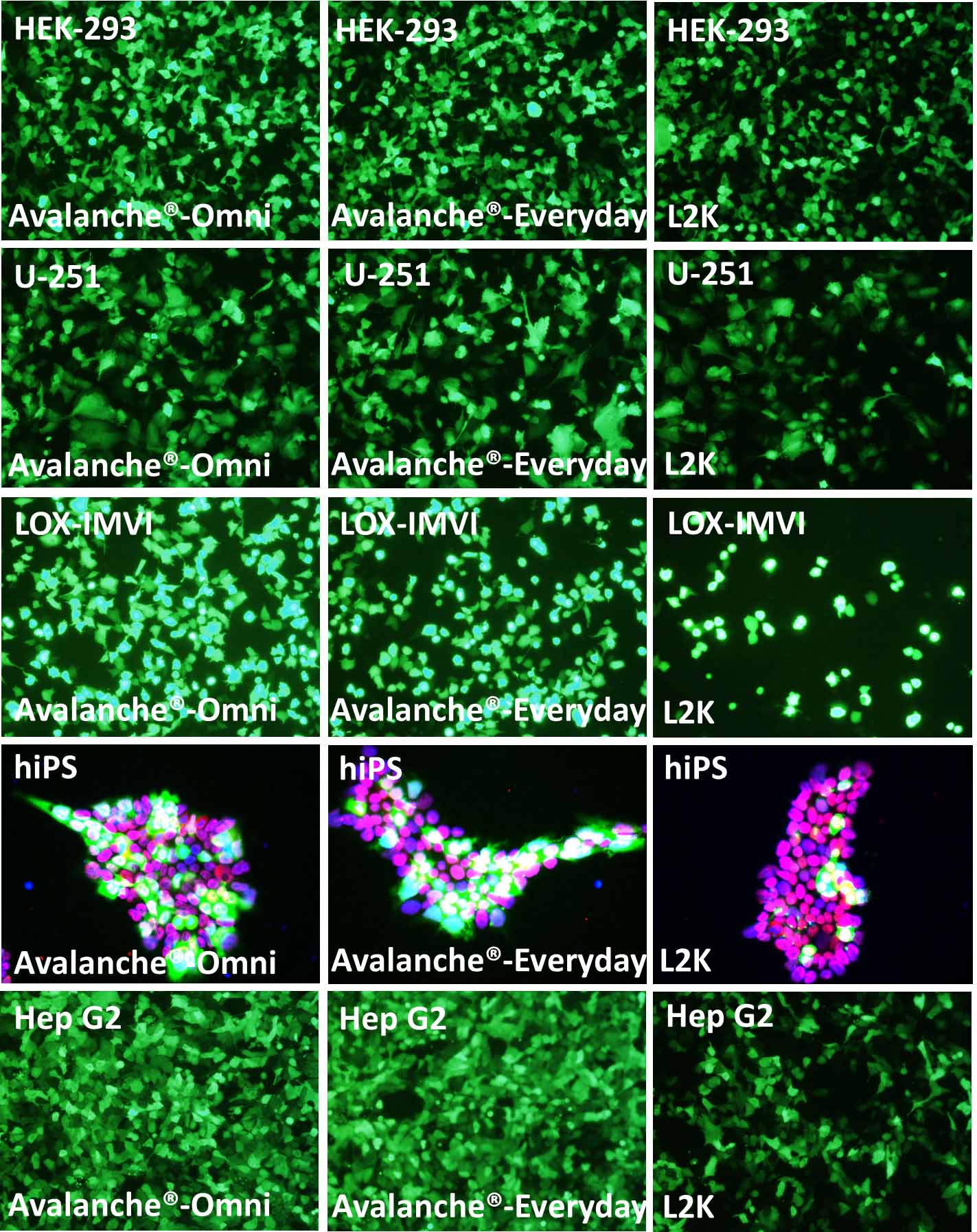 Figure 1. The performance of Avalanche®-Everyday Transfection Reagent (Avalanche®-Everyday) is very close to that of Avalanche®-Omni Transfection Reagent (Avalanche®-Omni). Cell line tested here are: HEK-293 (Human embryonic kidney cell line), U251 (Human glioblastoma cell), LOX-IMVI (Human melanoma cell), hiPS (Human induced pluripotent cell), and Hep G2 (Human hepatocellular carcinoma cell). The cells were transfected with enhanced green fluorescent protein (eGFP) expressing vector by using Avalanche®-Omni, Avalanche®-Everyday, and L2K. Regarding to hiPS cells, after transfection, hiPS cells were fixed with PFA, and stained with OCT-4 antibody (red) and DAPI (Blue). The pictures, which were the overlay of eGFP (green), Oct-4 (red), and DAPI (blue), showed that while all of the hiPS kept their pluripotency after transfection, Avalanche®-Omni and Avalanche®-Everyday showed great eGFP expression.
Figure 1. The performance of Avalanche®-Everyday Transfection Reagent (Avalanche®-Everyday) is very close to that of Avalanche®-Omni Transfection Reagent (Avalanche®-Omni). Cell line tested here are: HEK-293 (Human embryonic kidney cell line), U251 (Human glioblastoma cell), LOX-IMVI (Human melanoma cell), hiPS (Human induced pluripotent cell), and Hep G2 (Human hepatocellular carcinoma cell). The cells were transfected with enhanced green fluorescent protein (eGFP) expressing vector by using Avalanche®-Omni, Avalanche®-Everyday, and L2K. Regarding to hiPS cells, after transfection, hiPS cells were fixed with PFA, and stained with OCT-4 antibody (red) and DAPI (Blue). The pictures, which were the overlay of eGFP (green), Oct-4 (red), and DAPI (blue), showed that while all of the hiPS kept their pluripotency after transfection, Avalanche®-Omni and Avalanche®-Everyday showed great eGFP expression.
Avalanche®-Everyday and Avalanche®-Omni are able to achieved more than 90% knockdown of endogenous gene expression in a variety of cell lines and primary cells.
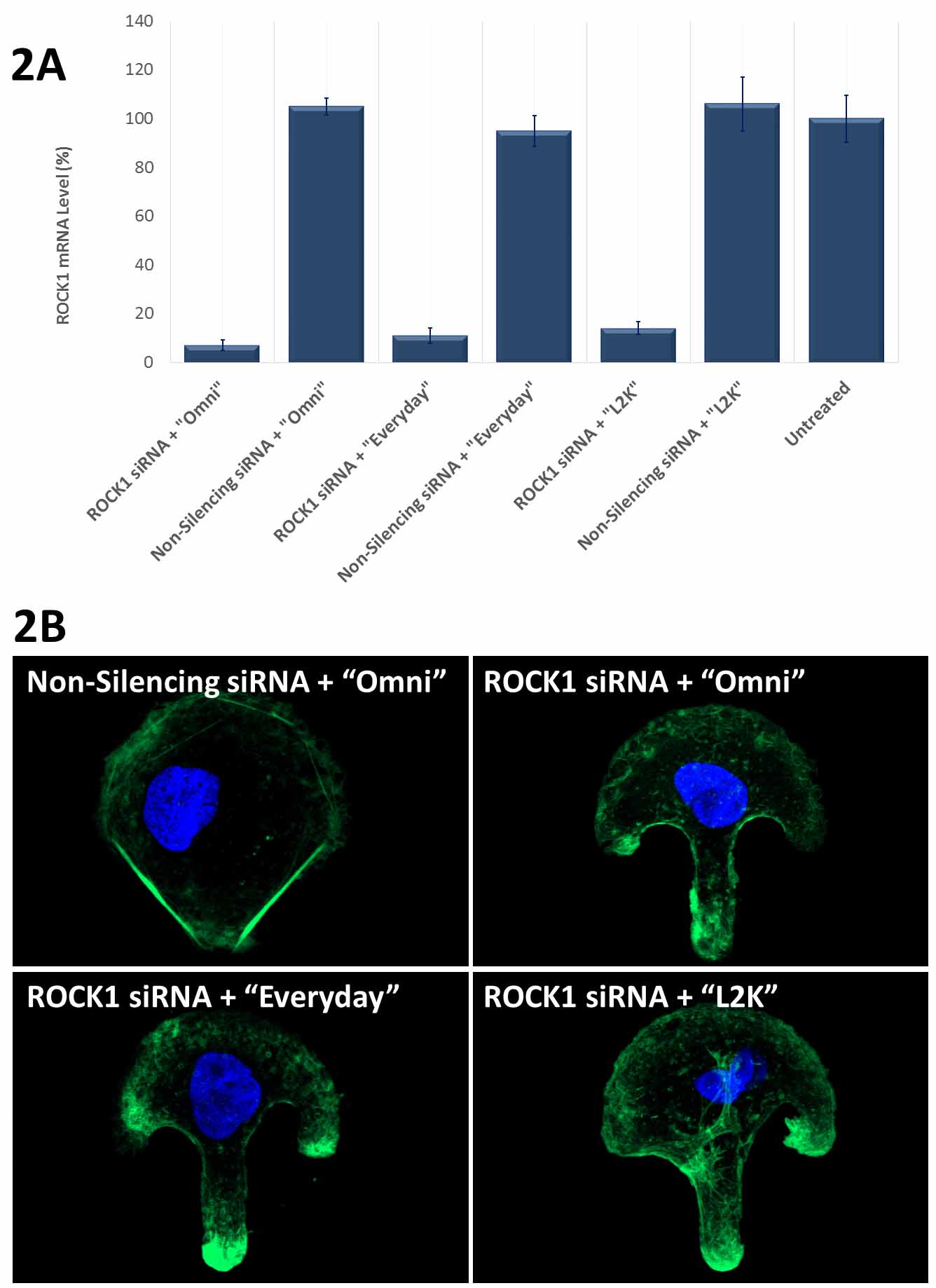 Figure 2. Avalanche®-Everyday Transfection Reagent (“Everyday”) and Avalanche®-Omni Transfection Reagent (“Omni”) achieved more than 90% knockdown of endogenous gene expression. (2A). ROCK1 mRNA levels were quantified using qPCR in Hela cells transfected with Target-specific siRNA duplexes (10nM) for human ROCK1 gene or non-silencing siRNA by using “Omni”, “Everyday”, and Lipofectamine® 2000 (“L2K”). Data were normalized against the 18S rRNA signal. Control Samples were either mock-transected or untreated. Values are normalized to untreated sample. Data are means®®SD (n=3). (B) Actin stress fibers were stained with FITC-labelled phalloidin on the cells in (A) cultured on fibronectin-coated micropattern. Confocal Microscope revealed that the cells transfected with ROCK1 siRNA showed disrupted stress fiber pattern as compared to the cells transfected with non-silencing siRNA using the same transfection reagent “Omni”. The use of “Everyday” showed similar disrupted stress fiber pattern as that of “Omni”, while the use of “L2K” only showed slightly disrupted stress fiber pattern. Lipofectamine® is a trademark of Life Technologies Corporation. Avalanche® is a trademark of EZ Biosystems® LLC
Figure 2. Avalanche®-Everyday Transfection Reagent (“Everyday”) and Avalanche®-Omni Transfection Reagent (“Omni”) achieved more than 90% knockdown of endogenous gene expression. (2A). ROCK1 mRNA levels were quantified using qPCR in Hela cells transfected with Target-specific siRNA duplexes (10nM) for human ROCK1 gene or non-silencing siRNA by using “Omni”, “Everyday”, and Lipofectamine® 2000 (“L2K”). Data were normalized against the 18S rRNA signal. Control Samples were either mock-transected or untreated. Values are normalized to untreated sample. Data are means®®SD (n=3). (B) Actin stress fibers were stained with FITC-labelled phalloidin on the cells in (A) cultured on fibronectin-coated micropattern. Confocal Microscope revealed that the cells transfected with ROCK1 siRNA showed disrupted stress fiber pattern as compared to the cells transfected with non-silencing siRNA using the same transfection reagent “Omni”. The use of “Everyday” showed similar disrupted stress fiber pattern as that of “Omni”, while the use of “L2K” only showed slightly disrupted stress fiber pattern. Lipofectamine® is a trademark of Life Technologies Corporation. Avalanche® is a trademark of EZ Biosystems® LLC
Additional Information
| Weight | 0.5 lbs |
|---|---|
| Product Sizes | 0.75 ml, 1.5 ml |
| Subcategories | Broad Spectrum |
Documents
Protocols
MSDS
Citations or Feedback
- Aso, S., Kitao, K., Hashimoto-Gotoh, A., Sakaguchi, S., & Miyazawa, T. (2021). Identification of Feline Foamy Virus-derived MicroRNAs. Microbes Environ, 36(4). doi:10.1264/jsme2.ME21055
- Ebihara, T., Masuda, A., Takahashi, D., Hino, M., Mon, H., Kakino, K., . . . Kusakabe, T. (2021). Production of scFv, Fab, and IgG of CR3022 Antibodies Against SARS-CoV-2 Using Silkworm-Baculovirus Expression System. Mol Biotechnol, 63(12), 1223-1234. doi:10.1007/s12033-021-00373-0
- Filippov-Levy, N., Davidson, B., & Reich, R. (2020). The Biological Role of the Long Non-coding RNA LINK-A in Ovarian Carcinoma. Anticancer Res, 40(12), 6677-6684. doi:10.21873/anticanres.14691
- Filippov-Levy, N., Reich, R., & Davidson, B. (2020). The Biological and Clinical Role of the Long Non-Coding RNA LOC642852 in Ovarian Carcinoma. Int J Mol Sci, 21(15). doi:10.3390/ijms21155237
- Fujita, R., Hino, M., Ebihara, T., Nagasato, T., Masuda, A., Lee, J. M., . . . Kusakabe, T. (2020). Efficient production of recombinant SARS-CoV-2 spike protein using the baculovirus-silkworm system. Biochem Biophys Res Commun, 529(2), 257-262. doi:10.1016/j.bbrc.2020.06.020
- Fujiwara, S., Nguyen, T. P., Furuse, K., Fukazawa, Y., Otani, T., & Furuse, M. (2022). Tight junction formation by a claudin mutant lacking the COOH-terminal PDZ domain-binding motif. Ann N Y Acad Sci, 1516(1), 85-94. doi:10.1111/nyas.14881
- Funato, N., & Yanagisawa, H. (2022). TBX1 targets the miR-200-ZEB2 axis to induce epithelial differentiation and inhibit stem cell properties. Sci Rep, 12(1), 20188. doi:10.1038/s41598-022-24604-9
- Garcia, B. C. B., Mukai, Y., Tomonaga, K., & Horie, M. (2023). The hidden diversity of ancient bornaviral sequences from X and P genes in vertebrate genomes. Virus Evol, 9(1), vead038. doi:10.1093/ve/vead038
- Gershanov, S., Toledano, H., Pernicone, N., Fichman, S., Michowiz, S., Pinhasov, A., . . . Salmon-Divon, M. (2021). Differences in RNA and microRNA Expression Between PTCH1- and SUFU-mutated Medulloblastoma. Cancer Genomics Proteomics, 18(3), 335-347. doi:10.21873/cgp.20264
- Hayashi, C., Takagi, K., Sato, A., Yamaguchi, M., Minemura, H., Miki, Y., . . . Suzuki, T. (2021). D-2-hydroxyglutarate dehydrogenase in breast carcinoma as a potent prognostic marker associated with proliferation. Histol Histopathol, 36(10), 1053-1062. doi:10.14670/HH-18-362
- Hernandez-Vicens, R., Singh, J., Pernicone, N., Listovsky, T., & Gerlitz, G. (2022). SETDB1 regulates microtubule dynamics. Cell Prolif, 55(12), e13348. doi:10.1111/cpr.13348
- Hino, M., Tatsuke, T., Morio, A., Mon, H., Lee, J. M., Masuda, A., . . . Kusakabe, T. (2022). Characterization of a Novel Heterochromatin Protein 1 Homolog “HP1c” in the Silkworm, Bombyx mori. Insects, 13(7). doi:10.3390/insects13070631
- Hirai, Y., & Horie, M. (2023). Nyamanini Virus Nucleoprotein and Phosphoprotein Organize Viral Inclusion Bodies That Associate with Host Biomolecular Condensates in the Nucleus. Int J Mol Sci, 24(7). doi:10.3390/ijms24076550
- Kamikawa, Y., Saito, A., Matsuhisa, K., Kaneko, M., Asada, R., Horikoshi, Y., . . . Imaizumi, K. (2021). OASIS/CREB3L1 is a factor that responds to nuclear envelope stress. Cell Death Discov, 7(1), 152. doi:10.1038/s41420-021-00540-x
- Kitao, K., Nakagawa, S., & Miyazawa, T. (2021). An ancient retroviral RNA element hidden in mammalian genomes and its involvement in co-opted retroviral gene regulation. Retrovirology, 18(1), 36. doi:10.1186/s12977-021-00580-2
- Kitao, K., Shoji, H., Miyazawa, T., & Nakagawa, S. (2023). Dynamic Evolution of Retroviral Envelope Genes in Egg-Laying Mammalian Genomes. Mol Biol Evol, 40(5). doi:10.1093/molbev/msad090
- Kitao, K., Tanikaga, T., & Miyazawa, T. (2019). Identification of a post-transcriptional regulatory element in the human endogenous retroviral syncytin-1. J Gen Virol, 100(4), 662-668. doi:10.1099/jgv.0.001238
- Kudo, T., Lazarou, M., Sliter, D. A., Kane, L. A., Sarraf, S. A., Wang, C., . . . Youle, R. J. (2021). The chicken homolog of KIAA0319L functions as a receptor of avian adeno-associated virus (A3V). The Shujitsu University journal of pharmaceutical sciences, 8(7565), 37. doi:10.1038/nature14893
- Mon, H., Sato, M., Lee, J. M., & Kusakabe, T. (2022). Construction of gene co-expression networks in cultured silkworm cells and identification of previously uncharacterized lepidopteran-specific genes required for chromosome dynamics. Insect Biochem Mol Biol, 151, 103875. doi:10.1016/j.ibmb.2022.103875
- Morio, A., Lee, J. M., Fujii, T., Mon, H., Masuda, A., Kakino, K., . . . Kusakabe, T. (2023). The biological role of core 1beta1-3galactosyltransferase (T-synthase) in mucin-type O-glycosylation in Silkworm, Bombyx mori. Insect Biochem Mol Biol, 156, 103936. doi:10.1016/j.ibmb.2023.103936
- Nakai, N., Sato, K., Tani, T., Saito, K., Sato, F., & Terada, S. (2019). Genetically encoded orientation probes for F-actin for fluorescence polarization microscopy. Microscopy (Oxf), 68(5), 359-368. doi:10.1093/jmicro/dfz022
- Ohga, H., Ito, K., Kakino, K., Mon, H., Kusakabe, T., Lee, J. M., & Matsuyama, M. (2021). Leptin Is an Important Endocrine Player That Directly Activates Gonadotropic Cells in Teleost Fish, Chub Mackerel. Cells, 10(12). doi:10.3390/cells10123505
- Onallah, H., Mannully, S. T., Davidson, B., & Reich, R. (2022). Exosome Secretion and Epithelial-Mesenchymal Transition in Ovarian Cancer Are Regulated by Phospholipase D. Int J Mol Sci, 23(21). doi:10.3390/ijms232113286
- Pernicone, N., Elias, M., Onn, I., Tobi, D., & Listovsky, T. (2022). Disrupting the MAD2L2-Rev1 Complex Enhances Cell Death upon DNA Damage. Molecules, 27(3). doi:10.3390/molecules27030636
- Pernicone, N., Grinshpon, S., & Listovsky, T. (2020). CDH1 binds MAD2L2 in a Rev1-like pattern. Biochem Biophys Res Commun, 531(4), 566-572. doi:10.1016/j.bbrc.2020.07.118
- Pernicone, N., Peretz, L., Grinshpon, S., & Listovsky, T. (2020). MDA-MB-157 Cell Line Presents High Levels of MAD2L2 and Dysregulated Mitosis. Anticancer Res, 40(10), 5471-5480. doi:10.21873/anticanres.14558
- Saito, A., Kamikawa, Y., Ito, T., Matsuhisa, K., Kaneko, M., Okamoto, T., . . . Imaizumi, K. (2023). p53-independent tumor suppression by cell-cycle arrest via CREB/ATF transcription factor OASIS. Cell Rep, 42(5), 112479. doi:10.1016/j.celrep.2023.112479
- Sato, A., Takagi, K., Yoshimura, A., Tsukamoto, W., Yamaguchi-Tanaka, M., Miki, Y., . . . Suzuki, T. (2023). Kallikrein-Related Peptidase 12 (KLK12) in Breast Cancer as a Favorable Prognostic Marker. Int J Mol Sci, 24(9). doi:10.3390/ijms24098419
- Shoji, H., Kitao, K., Miyazawa, T., & Nakagawa, S. (2023). Potentially reduced fusogenicity of syncytin-2 in New World monkeys. FEBS Open Bio, 13(3), 459-467. doi:10.1002/2211-5463.13555
- Shoji, H., Tsukasa, Y., Kitao, K., & Miyazawa, T. (2023). Characterization of ferret Pit1 as a receptor of feline leukemia virus subgroup B. J Vet Med Sci, 85(3), 326-328. doi:10.1292/jvms.22-0526
- Sugimoto, H., Fukuda, S., Yokawa, S., Hori, M., Ninomiya, H., Sato, T., . . . Suzuki, T. (2022). Visualization of osteocalcin and bone morphogenetic protein 2 (BMP2) secretion from osteoblastic cells by bioluminescence imaging. Biochem Biophys Res Commun, 635, 203-209. doi:10.1016/j.bbrc.2022.10.042
- Sumiyoshi, A., Kitao, K., & Miyazawa, T. (2022). Genetic and biological characterization of feline foamy virus isolated from a leopard cat (Prionailurus bengalensis) in Vietnam. J Vet Med Sci, 84(1), 157-165. doi:10.1292/jvms.21-0550
- Takagi, K., Miki, Y., Onodera, Y., Ishida, T., Watanabe, M., Sasano, H., & Suzuki, T. (2018). ARHGAP15 in Human Breast Carcinoma: A Potent Tumor Suppressor Regulated by Androgens. Int J Mol Sci, 19(3). doi:10.3390/ijms19030804
- Tanaka, M., Fujii, T., Mon, H., Lee, J. M., Kakino, K., Fukumori, H., . . . Kusakabe, T. (2022). Silkworm FoxL21 plays important roles as a regulator of ovarian development in both oogenesis and ovariole development. Insect Biochem Mol Biol, 143, 103737. doi:10.1016/j.ibmb.2022.103737
- Tawarayama, H., Feng, Q., Murayama, N., Suzuki, N., & Nakazawa, T. (2019). Cyclin-Dependent Kinase Inhibitor 2b Mediates Excitotoxicity-Induced Death of Retinal Ganglion Cells. Invest Ophthalmol Vis Sci, 60(13), 4479-4488. doi:10.1167/iovs.19-27396
- Terasawa, K., Kato, Y., Ikami, Y., Sakamoto, K., Ohtake, K., Kusano, S., . . . Hara-Yokoyama, M. (2021). Direct homophilic interaction of LAMP2A with the two-domain architecture revealed by site-directed photo-crosslinks and steric hindrances in mammalian cells. Autophagy, 17(12), 4286-4304. doi:10.1080/15548627.2021.1911017
- Tsujioka, S., Sumino, A., Nagasawa, Y., Sumikama, T., Flechsig, H., Puppulin, L., . . . Shibata, M. (2023). Imaging single CaMKII holoenzymes at work by high-speed atomic force microscopy. Sci Adv, 9(26), eadh1069. doi:10.1126/sciadv.adh1069
- Wang, X., Pernicone, N., Pertz, L., Hua, D., Zhang, T., Listovsky, T., & Xie, W. (2019). REV7 has a dynamic adaptor region to accommodate small GTPase RAN/Shigella IpaB ligands, and its activity is regulated by the RanGTP/GDP switch. J Biol Chem, 294(43), 15733-15742. doi:10.1074/jbc.RA119.010123
- Zheng, P., Obara, C. J., Szczesna, E., Nixon-Abell, J., Mahalingan, K. K., Roll-Mecak, A., . . . Blackstone, C. (2022). ER proteins decipher the tubulin code to regulate organelle distribution. Nature, 601(7891), 132-138. doi:10.1038/s41586-021-04204-9 nulla pariatur?”


